
To make it compile, I removed the -Werror=all . In Lib/MPM/MpmSimulationBase.cpp, I replaced all lines "if constexpr (!USE_APIC_BLEND_RPIC)" with "if (!USE_APIC_BLEND_RPIC)"; "if constexpr (USE_APIC_BLEND_RPIC)" with "if (USE_APIC_BLEND_RPIC)"; "if constexpr (!USE_MPM_DEGREE_ONE)" with "if (!USE_MPM_DEGREE_ONE)".
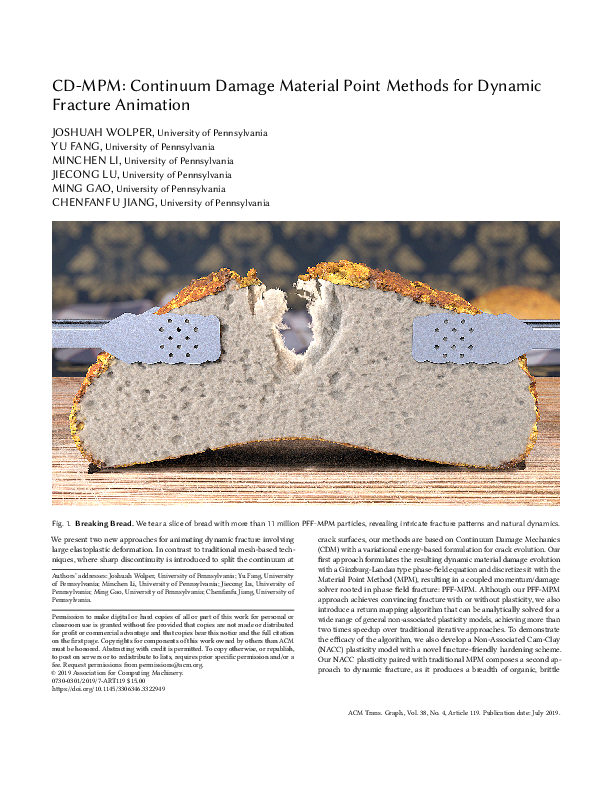
The code corresponds to two siggraph 2019 papers. It is not explicit which code corresponds to which paper, I assumed "fracture" corresponds to this paper. I could reproduce the dino and bread experiments. I visualized the bgeo files with the partview tool available here: "https://github.com/wdas/partio", it can also be done using Houdini. To know which test number corresponds to which experiment, refer to the FractureInit3D.h file.
Compiles out of the box.
The code corresponds to two siggraph 2019 papers. It is not explicit which code corresponds to which paper, I assumed "fracture" corresponds to this paper. I could reproduce the dino and bread experiments. I visualized the bgeos files with the partview tool available here: "https://github.com/wdas/partio", it can also be done using Houdini. To know which test number corresponds to which experiment, refer to the FractureInit3D.h file.
If you want to contribute with another review, please follow these instructions.
Please consider to cut/paste/edit the raw JSON data attached to this paper.